flowchart LR
A1[("Training Data (X and y)")] --> B{{fit}}
A2[Model Object] --> B
B --> FM[Fitted Model]
FM --> C{{Predict}}
B2[(New data X)] --> C
C --> D[("predicted y's")]
Introduction to Supervised Learning
Day 1: Introduction
Think of examples
How is data used to make decisions that affect you or your neighbors?
Use whatever resources you like.
Predictive Analytics
- A powerful tool to turn data into action.
- It works because God made the universe predictable (and successful prediction rewarding), and promised order:
- covenant with Noah that the seasons would continue (Gen 8:22)
- guarantee in Psalms that the sun would rise (Ps 104:19)
- promise to Jeremiah that the sun, moon, and stars would continue (Jer 31:35-36)
- Need for wisdom: It can be used for great good and great harm
Power of Predictive Modeling
- Medicine: wearable monitor for seizures or falls, detect malaria from blood smears, find effective drug regimens from medical records
- Drug Discovery: predict the efficacy of a synthesis plan for a drug
- Precision Agriculture: predict effect of micro-climate on plant growth
- Urban Planning: forecast resource needs, extreme weather risks, …
- Government: classify feedback from constituents
- Retail: predict items in a grocery order
- Recommendation systems: Amazon, Netflix, YouTube, …
- User interfaces: gesture typing, autocomplete / autocorrect
and so much more…
The universe is surprisingly predictable
- God created the world with actionable structure
- We gradually learn how to perceive that structure and act within it.
- The better our perceptions align with how the universe is structured, the better our actions
- We can discover that structure by learning to be less surprised by what we see ( = predicting our perceptions)
- Perceptions are thus both accurate and fallable.
Predictive modeling technology: Need for wisdom
- Potential for great good, but also great harm:
- Lack of fairness in facial recognition, criminal risk, lending, job applicant scoring, …
- Lack of transparency in how “Big Data” systems make conclusions
- Lack of privacy as data is increasingly collected and aggregated
- Amplification of extreme positions in social media, YouTube, etc.
- Oversimplification of human experience
- Hidden human labor
- Illusion of objectivity
- …!
See, for example, Weapons of Math Destruction
Reminders from Statistics
- We might wish we had all possible data…
- We must acknowledge our uncertainty.
- Our measurements are partial.
- Our inferences sometimes fail (and we may not know it!)
- But God made a world with structure that we can learn about even with imperfect tools.
Activity
Teachable Machine
Pick a partner (or two). Decide who’s trainer and who’s tester.
Trainer (needs a laptop):
- Go to https://teachablemachine.withgoogle.com/train/image
- Train a simple classifier using your webcam. Ideas:
- which hand are you holding up? happy or sad face? looking left or right? two colors?
- Don’t worry about making it super accurate.
Tester:
- Try out your partner’s classifier (without touching the computer or the same objects).
- Try to find some situations where it works
- Try to write a succinct description of what situations cause it to behave unexpectedly.
We’ll share descriptions.
Supervised Learning and the scikit-learn API
Scikit-Learn (sklearn)
- A Python library for machine learning
- Provides a consistent API for many different kinds of models
- Provides many tools for data preparation, model evaluation, etc.
Model API: fit, predict
Example: penguins
import pandas as pd
penguins = pd.read_csv("https://raw.githubusercontent.com/allisonhorst/palmerpenguins/main/inst/extdata/penguins.csv")
penguins.info()<class 'pandas.core.frame.DataFrame'>
RangeIndex: 344 entries, 0 to 343
Data columns (total 8 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 species 344 non-null object
1 island 344 non-null object
2 bill_length_mm 342 non-null float64
3 bill_depth_mm 342 non-null float64
4 flipper_length_mm 342 non-null float64
5 body_mass_g 342 non-null float64
6 sex 333 non-null object
7 year 344 non-null int64
dtypes: float64(4), int64(1), object(3)
memory usage: 21.6+ KBWhich species is each penguin?
from sklearn.neighbors import KNeighborsClassifier
model = KNeighborsClassifier(n_neighbors=1).fit(
X=penguins_simple[['bill_depth_mm', 'bill_length_mm']],
y=penguins_simple['species'])
penguins_simple['predicted_species'] = model.predict(
X=penguins_simple[['bill_depth_mm', 'bill_length_mm']])
penguins_simple.head()| bill_depth_mm | bill_length_mm | species | predicted_species | |
|---|---|---|---|---|
| 0 | 18.7 | 39.1 | Adelie | Adelie |
| 1 | 17.4 | 39.5 | Adelie | Adelie |
| 2 | 18.0 | 40.3 | Adelie | Adelie |
| 4 | 19.3 | 36.7 | Adelie | Adelie |
| 5 | 20.6 | 39.3 | Adelie | Adelie |
Are those predictions are good?
| species | predicted_species | size | |
|---|---|---|---|
| 0 | Adelie | Adelie | 151 |
| 1 | Chinstrap | Chinstrap | 68 |
| 2 | Gentoo | Gentoo | 123 |
Well, of course…
Why should we be unsurprised that this works?
Train-test split!
flowchart LR
A1[("All training data")] --> B{{Split}}
B --> A2[("Training data")]
B --> A3[("Test data")]
A2 --> C{{Fit}}
A3 -->|NEVER!| C
C --> D[("Fitted model")]
import numpy as np
np.random.seed(0)
penguins_simple["use_for_training"] = np.random.rand(len(penguins_simple)) < 0.5
penguins_train = penguins_simple.query("use_for_training").copy()
penguins_test = penguins_simple.query("not use_for_training").copy()
print(f"{len(penguins_train)} training penguins, {len(penguins_test)} test penguins")171 training penguins, 171 test penguinsNote: Later we’ll learn about the train_test_split function, which does this for us.
Train on the training set
How does it do?
penguins_train['predicted_species'] = model.predict(
X=penguins_train[['bill_depth_mm', 'bill_length_mm']])
penguins_train.groupby(['species', 'predicted_species'], as_index=False).size()| species | predicted_species | size | |
|---|---|---|---|
| 0 | Adelie | Adelie | 69 |
| 1 | Chinstrap | Chinstrap | 39 |
| 2 | Gentoo | Gentoo | 63 |
penguins_test['predicted_species'] = model.predict(
X=penguins_test[['bill_depth_mm', 'bill_length_mm']])
penguins_test.groupby(['species', 'predicted_species'], as_index=False).size()| species | predicted_species | size | |
|---|---|---|---|
| 0 | Adelie | Adelie | 81 |
| 1 | Adelie | Chinstrap | 1 |
| 2 | Chinstrap | Adelie | 2 |
| 3 | Chinstrap | Chinstrap | 26 |
| 4 | Chinstrap | Gentoo | 1 |
| 5 | Gentoo | Chinstrap | 6 |
| 6 | Gentoo | Gentoo | 54 |
Other kinds of models
- Nearest Neighbors (
KNeighborsClassifier) - Decision Trees (
DecisionTreeClassifier) and Random Forests (RandomForestClassifier) - Linear Models (
LogisticRegression, yes it’s a classifier) - Neural Networks (
MLPClassifier) - Boosting methods (
HistGradientBoostingClassifier)
Many models, same API
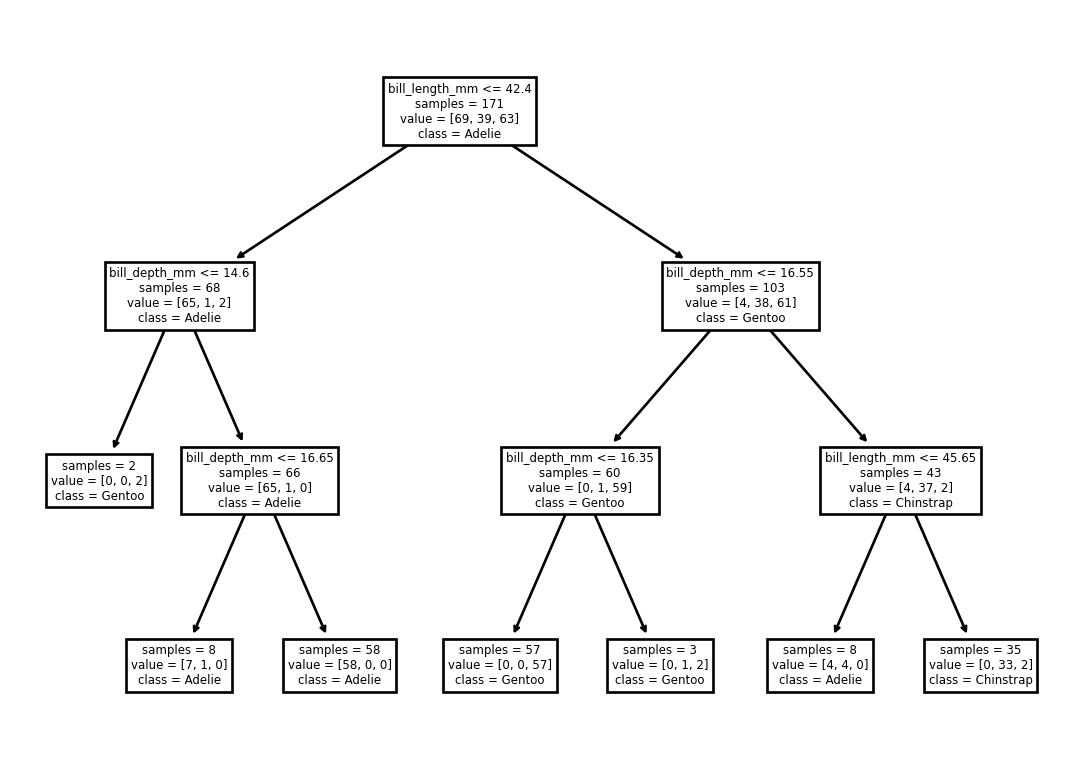
Decision tree:
from sklearn.tree import DecisionTreeClassifier, plot_tree
model = DecisionTreeClassifier(max_depth=3).fit(
X=penguins_train[['bill_depth_mm', 'bill_length_mm']],
y=penguins_train['species'])
model.score(
X=penguins_test[['bill_depth_mm', 'bill_length_mm']],
y=penguins_test['species'])0.9122807017543859What does that tree do?
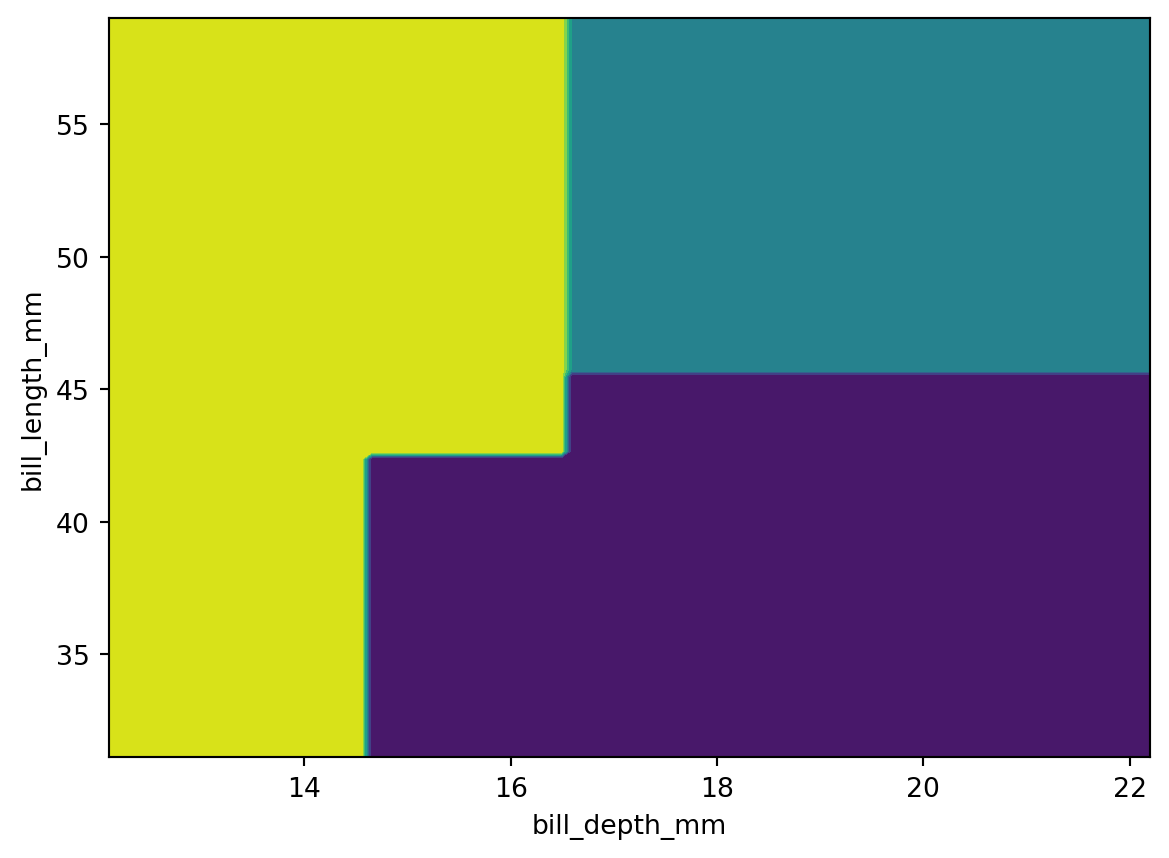
Linear Model (Logistic Regression)
array([[ 1.91703649, -0.89183601],
[ 0.39297577, 0.28951405],
[-2.31001227, 0.60232196]])Neural Network
from sklearn.neural_network import MLPClassifier
model = MLPClassifier(hidden_layer_sizes=(10, 10)).fit(
X=penguins_train[['bill_depth_mm', 'bill_length_mm']],
y=penguins_train['species'])/usr/local/Caskroom/miniconda/base/envs/fa22/lib/python3.10/site-packages/sklearn/neural_network/_multilayer_perceptron.py:702: ConvergenceWarning:
Stochastic Optimizer: Maximum iterations (200) reached and the optimization hasn't converged yet.
Wednesday Slides
Recall
- What does
fitdo? - What does
predictdo? - Why did we get an
accuracy_scoreof 1.0? What did we do wrong? - What should you never do with the test set?
Types of Learning from Data
- Supervised Learning: Filling in a label for each item based on features
- Classification: Labels are discrete (e.g. penguin species, spam/not spam, which word was swiped, …)
- Regression: Labels are continuous (e.g. sale price, seconds of video watched, …)
- Forecasting: predict how a sequence will continue (future observations)
- Unsupervised Learning: Finding relationships between items, doesn’t need labels
- Clustering: Grouping similar items (e.g. customer segments, …)
- Dimensionality Reduction: Finding a lower-dimensional representation of data (e.g. word embeddings, …)
We’ll focus on supervised learning and clustering in this course. Others:
- Reinforcement Learning: Learning to take actions to maximize a reward (e.g. game playing, robotics, …)
- Self-supervised learning: turning an unsupervised problem into a supervised one (e.g. predicting the next word in a sentence)
- Statistical inference: fill in summary statistics (we wanted a population but only got a sample)
- Causal inference: fill in counterfactuals (what if?)
Regression Example 1
Concrete Mixture Strength
From Applied Predictive Modeling (M. Kuhn and Johnson 2013), chapter 10.
Original column names:
['Cement (component 1)(kg in a m^3 mixture)', 'Blast Furnace Slag (component 2)(kg in a m^3 mixture)', 'Fly Ash (component 3)(kg in a m^3 mixture)', 'Water (component 4)(kg in a m^3 mixture)', 'Superplasticizer (component 5)(kg in a m^3 mixture)', 'Coarse Aggregate (component 6)(kg in a m^3 mixture)', 'Fine Aggregate (component 7)(kg in a m^3 mixture)', 'Age (day)', 'Concrete compressive strength(MPa. megapascals)']# clean up column names: remove everything after the first parenthesis, replace spaces with underscores
concrete_all.columns = [
col.split('(', 1)[0].strip().replace(' ', '_').lower()
for col in concrete_all.columns]
concrete_all = concrete_all.rename(columns={
'concrete_compressive_strength': 'strength',
'blast_furnace_slag': 'slag',
})
concrete_all.info()<class 'pandas.core.frame.DataFrame'>
RangeIndex: 1030 entries, 0 to 1029
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 cement 1030 non-null float64
1 slag 1030 non-null float64
2 fly_ash 1030 non-null float64
3 water 1030 non-null float64
4 superplasticizer 1030 non-null float64
5 coarse_aggregate 1030 non-null float64
6 fine_aggregate 1030 non-null float64
7 age 1030 non-null float64
8 strength 1030 non-null float64
dtypes: float64(9)
memory usage: 72.5 KBConcrete Mixture Strength data
| cement | slag | fly_ash | water | superplasticizer | coarse_aggregate | fine_aggregate | age | strength | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 540.0 | 0.0 | 0.0 | 162.0 | 2.5 | 1040.0 | 676.0 | 28.0 | 79.99 |
| 1 | 540.0 | 0.0 | 0.0 | 162.0 | 2.5 | 1055.0 | 676.0 | 28.0 | 61.89 |
| 2 | 332.5 | 142.5 | 0.0 | 228.0 | 0.0 | 932.0 | 594.0 | 270.0 | 40.27 |
| 3 | 332.5 | 142.5 | 0.0 | 228.0 | 0.0 | 932.0 | 594.0 | 365.0 | 41.05 |
| 4 | 198.6 | 132.4 | 0.0 | 192.0 | 0.0 | 978.4 | 825.5 | 360.0 | 44.30 |
Modeling Workflow
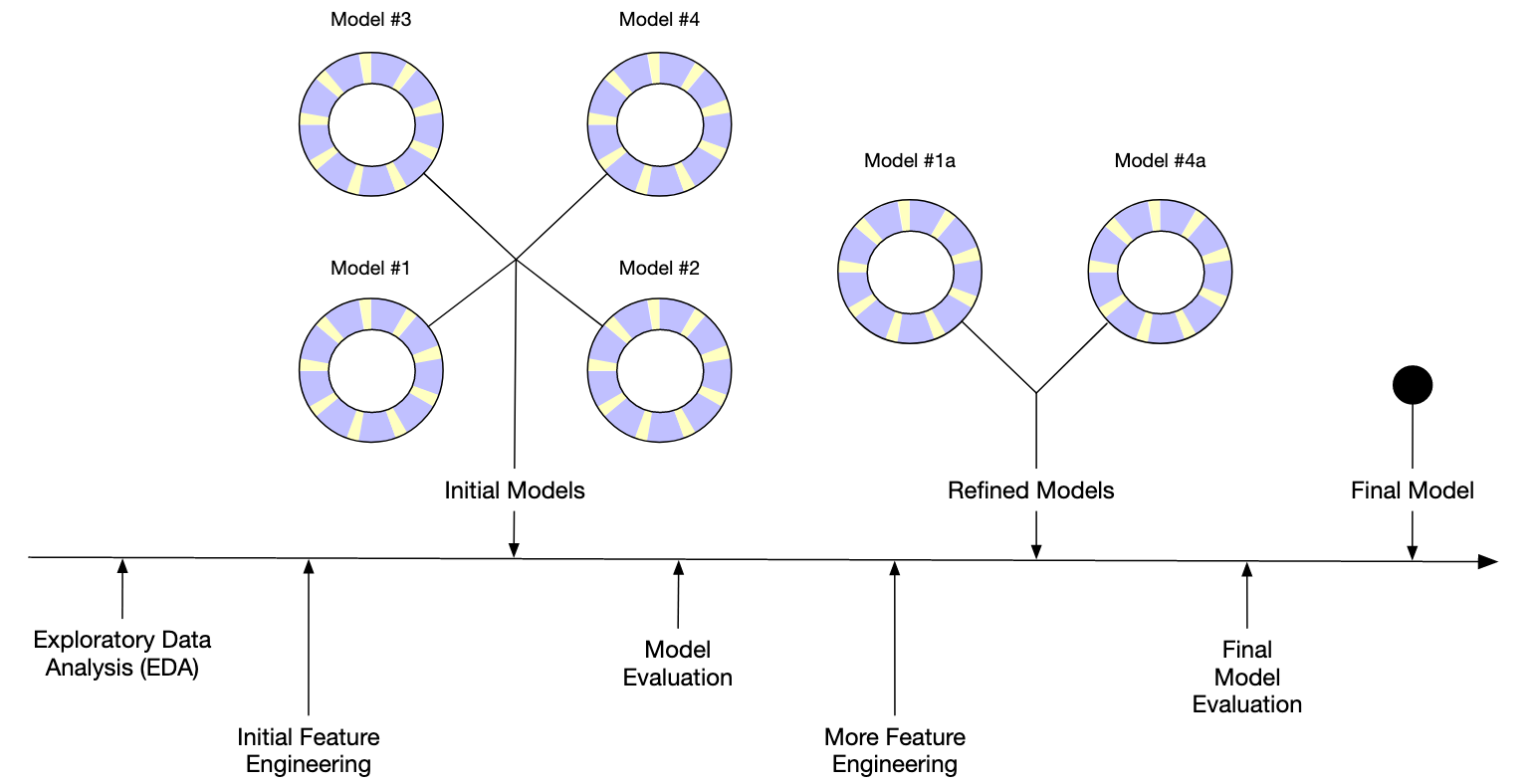
Step 1: Define the problem
- What are you trying to predict?
- terms: target, response, label
- What data do you have?
- terms: features, predictors, covariates
- What metrics will indicate success? (Measure success in multiple ways!)
- terms: loss, error, score, metric
- penguins example: accuracy (perhaps broken down by species)
Concrete example:
- Task: predict concrete strength (MPa) from mixture ingredients
- Target: concrete strength
- Features: mixture ingredients (cement, slag, ash, water, …)
- Metric: how close are the predictions to the actual strength?
Step 2: Explore the data
- What is the structure of the data?
- What are the distributions of the features and target?
- Anything unexpected? (e.g. missing values, outliers, …): make lots of plots!
Concrete example:
# Do we have duplicates of any mixtures that differ only in age and strength?
other_columns = [col for col in concrete_all.columns if col not in ['age', 'strength']]
concrete_all.groupby(other_columns, as_index=False).size().sort_values(by='size', ascending=False).head(10)| cement | slag | fly_ash | water | superplasticizer | coarse_aggregate | fine_aggregate | size | |
|---|---|---|---|---|---|---|---|---|
| 356 | 362.6 | 189.0 | 0.0 | 164.9 | 11.6 | 944.7 | 755.8 | 20 |
| 393 | 425.0 | 106.3 | 0.0 | 153.5 | 16.5 | 852.1 | 887.1 | 15 |
| 398 | 446.0 | 24.0 | 79.0 | 162.0 | 11.6 | 967.0 | 712.0 | 12 |
| 413 | 500.0 | 0.0 | 0.0 | 200.0 | 0.0 | 1125.0 | 613.0 | 8 |
| 355 | 359.0 | 19.0 | 141.0 | 154.0 | 10.9 | 942.0 | 801.0 | 8 |
| 426 | 540.0 | 0.0 | 0.0 | 173.0 | 0.0 | 1125.0 | 613.0 | 7 |
| 421 | 525.0 | 0.0 | 0.0 | 189.0 | 0.0 | 1125.0 | 613.0 | 7 |
| 342 | 339.0 | 0.0 | 0.0 | 197.0 | 0.0 | 968.0 | 781.0 | 7 |
| 367 | 380.0 | 95.0 | 0.0 | 228.0 | 0.0 | 932.0 | 594.0 | 6 |
| 147 | 198.6 | 132.4 | 0.0 | 192.0 | 0.0 | 978.4 | 825.5 | 6 |
We’ll need to address this later.
Are they all the same total weight?
Step 3: Split the data
So we can evaluate the model.

Splitting concrete data
from sklearn.model_selection import train_test_split
concrete_train, concrete_test = train_test_split(concrete_all, random_state=0, test_size=0.2)
print(f"{len(concrete_train)} training mixtures, {len(concrete_test)} test mixtures")824 training mixtures, 206 test mixturesCareful: we just duplicated some mixtures in both sets (measurements at different ages).
Step 4: Feature Engineering
Sometimes the features are already in a useful form, but sometimes we can get more useful features by transforming or combining the existing features.
Examples:
- ratios of ingredients (e.g., coarse aggregate / fine aggregate)
- log of
age(influence of a day decreases as age increases) - polynomial features (e.g. \(x^2\))
- product of two features (interactions)
- date features (e.g., day of week, month)
- replace rare categories with “other”
We’ll skip this for concrete for now.
Step 5: Train (fit) and evaluate some models
- Pick a model (or several) that’s appropriate for the task and data
- Fit the model to the training data
- Evaluate the model on the test data
Some models may require special preparation of the data:
- Missing values
- Categorical variables
- Scaling
- Feature engineering
Step 5: a linear model for concrete
from sklearn.linear_model import LinearRegression
feature_columns = [col for col in concrete_train.columns if col != 'strength']
model = LinearRegression().fit(
X=concrete_train[feature_columns],
y=concrete_train['strength'])
concrete_test['predicted_strength'] = model.predict(
X=concrete_test[feature_columns])Metrics: Were those predictions good?
from sklearn.metrics import mean_absolute_error, mean_absolute_percentage_error, mean_squared_error, r2_score
def evaluate(y_true, y_pred):
return pd.Series({
'MAE': mean_absolute_error(y_true, y_pred),
'MAPE': mean_absolute_percentage_error(y_true, y_pred),
'R^2': r2_score(y_true, y_pred),
})
def evaluate_df(df):
return evaluate(df['strength'], df['predicted_strength'])
evaluate_df(concrete_test).to_frame('test')| test | |
|---|---|
| MAE | 7.864642 |
| MAPE | 0.331604 |
| R^2 | 0.636961 |
- MAE: Mean Absolute Error (“predictions are usually off by xxx MPa”)
- MAPE: Mean Absolute Percent Error (“predictions are usually off by yy%”)
- Traditional R^2 (fraction of variance explained)
Note: Above, we defined a function to evaluate these errors on any model. That way we can evaluate many models in the same way.
Step 6: Tune the model
- Try different models and hyperparameters
- Try different feature engineering
- Analyze model errors (more EDA)
- Repeat steps 4-6 until model is good enough
EDA on model errors for Concrete
concrete_test['error'] = concrete_test['strength'] - concrete_test['predicted_strength']
px.histogram(concrete_test, x='error', nbins=30)Does the model do better or worse for certain ranges?
px.scatter(
concrete_test,
x='strength', y='error',
trendline='ols',
labels={
'strength': 'Strength (MPa)',
'error': 'Error (MPa)',
},
)concrete_test['strength_bin'] = pd.cut(concrete_test['strength'], bins=5)
concrete_test.groupby('strength_bin', as_index=False).apply(evaluate_df)| strength_bin | MAE | MAPE | R^2 | |
|---|---|---|---|---|
| 0 | (4.495, 19.516] | 9.000968 | 0.848740 | -5.671546 |
| 1 | (19.516, 34.462] | 5.808164 | 0.213878 | -2.287072 |
| 2 | (34.462, 49.408] | 8.305960 | 0.201023 | -5.271035 |
| 3 | (49.408, 64.354] | 8.740660 | 0.154203 | -4.768868 |
| 4 | (64.354, 79.3] | 11.762144 | 0.164826 | -7.339047 |
concrete_test['age_bin'] = pd.cut(concrete_test['age'], bins=3)
concrete_test.groupby('age_bin', as_index=False).apply(evaluate_df)| age_bin | MAE | MAPE | R^2 | |
|---|---|---|---|---|
| 0 | (2.638, 123.667] | 7.760751 | 0.339492 | 0.652625 |
| 1 | (123.667, 244.333] | 6.864034 | 0.188294 | 0.568809 |
| 2 | (244.333, 365.0] | 11.220562 | 0.268660 | -2.173470 |
Any relationships with other features?
We’re not seeing any clear overall patterns. Which mixtures are we worst at predicting? (ask a domain expert about these)
| cement | slag | fly_ash | water | superplasticizer | coarse_aggregate | fine_aggregate | age | strength | predicted_strength | error | strength_bin | age_bin | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 477 | 446.0 | 24.0 | 79.0 | 162.0 | 11.6 | 967.0 | 712.0 | 3.0 | 23.35 | 50.426023 | -27.076023 | (19.516, 34.462] | (2.638, 123.667] |
| 85 | 379.5 | 151.2 | 0.0 | 153.9 | 15.9 | 1134.3 | 605.0 | 3.0 | 28.60 | 52.987434 | -24.387434 | (19.516, 34.462] | (2.638, 123.667] |
| 622 | 307.0 | 0.0 | 0.0 | 193.0 | 0.0 | 968.0 | 812.0 | 365.0 | 36.15 | 59.850056 | -23.700056 | (34.462, 49.408] | (244.333, 365.0] |
| 604 | 339.0 | 0.0 | 0.0 | 197.0 | 0.0 | 968.0 | 781.0 | 365.0 | 38.89 | 62.387515 | -23.497515 | (34.462, 49.408] | (244.333, 365.0] |
| 294 | 168.9 | 42.2 | 124.3 | 158.3 | 10.8 | 1080.8 | 796.2 | 3.0 | 7.40 | 28.205935 | -20.805935 | (4.495, 19.516] | (2.638, 123.667] |
| cement | slag | fly_ash | water | superplasticizer | coarse_aggregate | fine_aggregate | age | strength | predicted_strength | error | strength_bin | age_bin | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 358 | 277.2 | 97.8 | 24.5 | 160.7 | 11.2 | 1061.7 | 782.5 | 100.0 | 66.95 | 48.291387 | 18.658613 | (64.354, 79.3] | (2.638, 123.667] |
| 828 | 522.0 | 0.0 | 0.0 | 146.0 | 0.0 | 896.0 | 896.0 | 28.0 | 74.99 | 53.777042 | 21.212958 | (64.354, 79.3] | (2.638, 123.667] |
| 356 | 277.2 | 97.8 | 24.5 | 160.7 | 11.2 | 1061.7 | 782.5 | 28.0 | 63.14 | 39.996732 | 23.143268 | (49.408, 64.354] | (2.638, 123.667] |
| 8 | 266.0 | 114.0 | 0.0 | 228.0 | 0.0 | 932.0 | 670.0 | 28.0 | 45.85 | 19.463945 | 26.386055 | (34.462, 49.408] | (2.638, 123.667] |
| 14 | 304.0 | 76.0 | 0.0 | 228.0 | 0.0 | 932.0 | 670.0 | 28.0 | 47.81 | 19.859987 | 27.950013 | (34.462, 49.408] | (2.638, 123.667] |
Concrete: try a different model
e.g., random forest
Your turn: how well does a Random Forest model do on the test set?
Step 7: Deploy the model
Often called “inference” (as opposed to “training”)
- Use the model to make predictions on new data as it arrives
- Monitor the model’s performance over time
- Retrain the model periodically