Trees
Imports …
Example: Blood test for autism?
We’ll use an example from a 2017 PLOS Computational Biology paper. Download the data.
- Typically autism is diagnosed by behavioral symptoms
- If we could diagnose autism from a blood test, we could diagnose it earlier
The data has units on the second row, so we’ll skip that row.
We have 3 kinds of data about 206 children:
- The outcome (
Group): ASD (diagnosed with ASD), SIB (sibling not diagnosed with ASD), and NEU (age-matched neurotypical children, for control)
- The outcome (
Group): ASD, SIB, NEU - Concentrations of various metabolites in a blood sample:
```{python}
#| output: asis
print('\n'.join(f'- {column_name}' for column_name in autism.columns[1:-1]))
```- Methion.
- SAM
- SAH
- SAM/SAH
- % DNA methylation
- 8-OHG
- Adenosine
- Homocysteine
- Cysteine
- Glu.-Cys.
- Cys.-Gly.
- tGSH
- fGSH
- GSSG
- fGSH/GSSG
- tGSH/GSSG
- Chlorotyrosine
- Nitrotyrosine
- Tyrosine
- Tryptophane
- fCystine
- fCysteine
- fCystine/fCysteine
- % oxidized
- The outcome (
Group): ASD, SIB, NEU - Concentrations of various metabolites in a blood sample
- For the ASD children only, a measure of life skills (“Vineland ABC”)
Exploratory Data Analysis (EDA)
What do these metabolites look like?
Exploratory Data Analysis (EDA)
EDA
- This plot helps us compare different metabolites within each group.
- Better question for predictive task: Which of these metabolites help us distinguish autism?
Approach:
- easier to compare within a plot than across a facet, so switch y variable to Group.
- absolute values don’t matter much, so let each metabolite have its own x scale.
- plotly boxplots don’t show up well when small, so switch to a ridgeline plot
Can we predict ASD vs non-ASD from metabolites?
- Let’s start by (1) ignoring the behavior scores (that’s an outcome) and comparing just ASD and NEU.
- We need to drop SIB.
Train-test split
First Model: guessing most common
What if we always guessed the most common outcome?
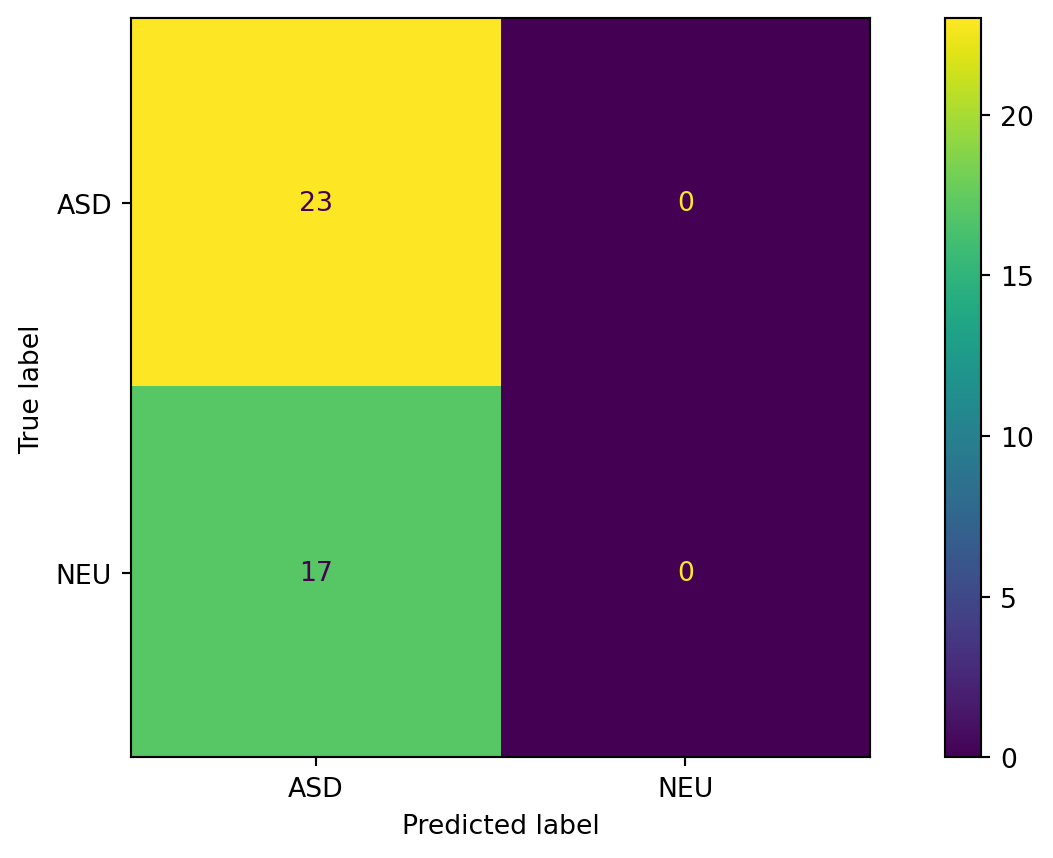
Confusion Matrix for most common
Uniform Random Guess
Or what if we guess uniformly at random?
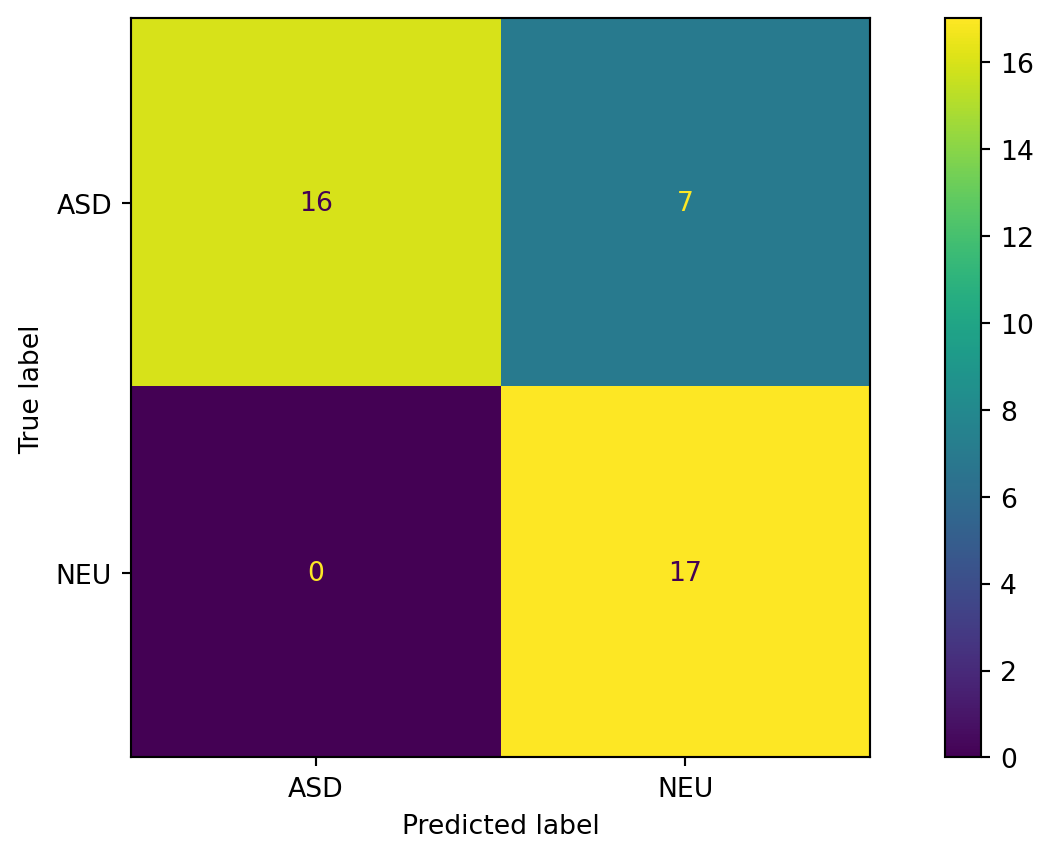
Confusion Matrix for Guessing Uniformly
Exercise: compute the following for this classifier:
- Accuracy
- False positive rate
- False negative rate
- Sensitivity
- Specificity
- Precision
- Recall
Computing these automatically
Now we can compute them (see docs on classification metrics)
from sklearn.metrics import classification_report
print(classification_report(
y_true=test[target_column],
y_pred=test["pred_uniform"],
)) precision recall f1-score support
ASD 0.65 0.48 0.55 23
NEU 0.48 0.65 0.55 17
accuracy 0.55 40
macro avg 0.56 0.56 0.55 40
weighted avg 0.58 0.55 0.55 40
Let’s compute these quantities straight from the confusion matrix.
from sklearn.metrics import confusion_matrix
(tn, fp), (fn, tp) = confusion_matrix(
y_true=test[target_column],
y_pred=test["pred_uniform"],
labels=[negative_outcome, positive_outcome]
)
num_positives = tp + fn
num_negatives = tn + fp
print(f"Accuracy: {(tp + tn) / (num_positives + num_negatives):.2f}")
print(f"False positive rate: {fp / num_negatives:.2f}")
print(f"False negative rate: {fn / num_positives:.2f}")
print(f"Sensitivity / recall: {tp / num_positives:.2f}")
print(f"Specificity: {tn / (tn + fp):.2f}")
print(f"Precision: {tp / (tp + fp):.2f}")Accuracy: 0.55
False positive rate: 0.35
False negative rate: 0.52
Sensitivity / recall: 0.48
Specificity: 0.65
Precision: 0.65Decision Tree
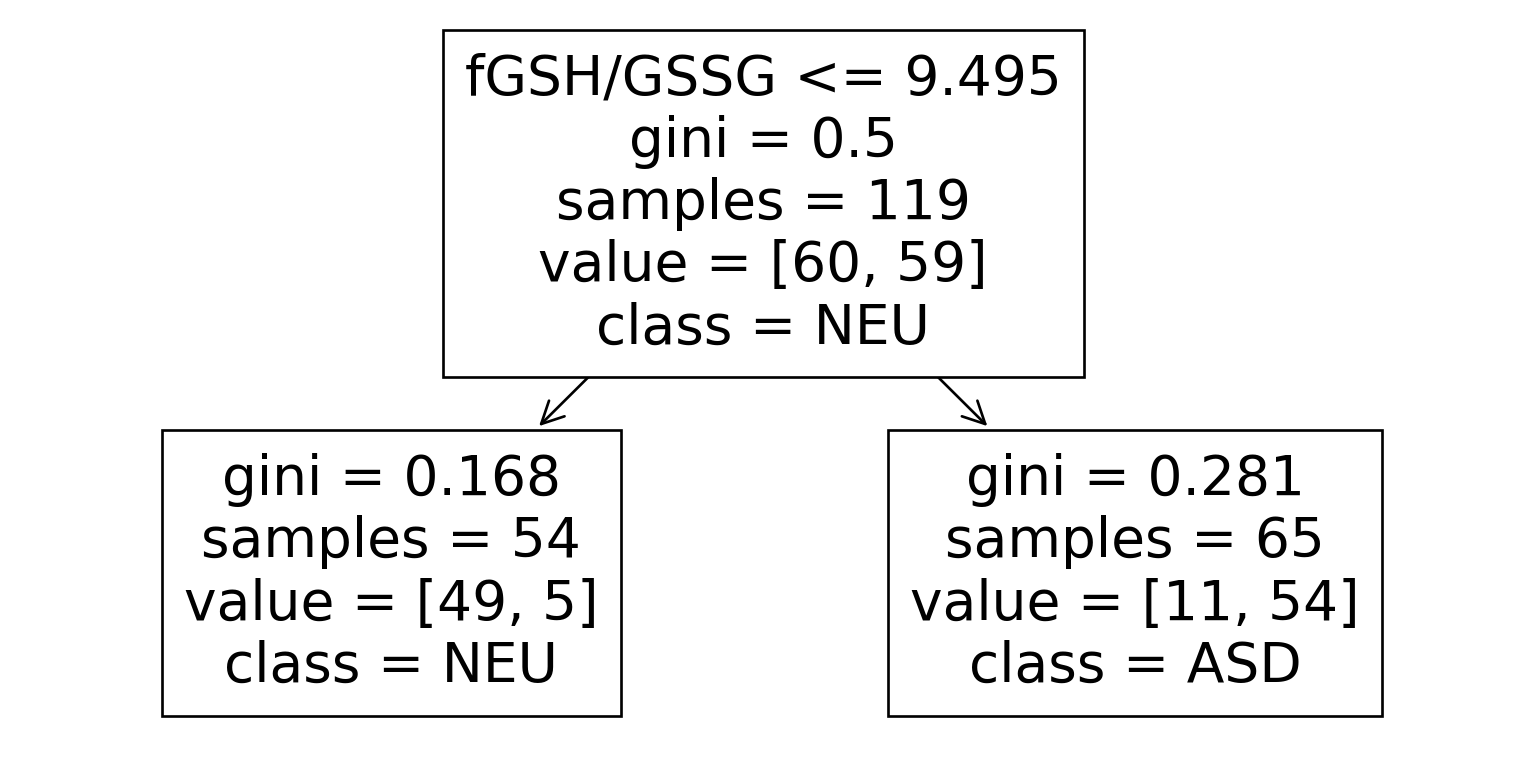
from sklearn.tree import DecisionTreeClassifier
tree = DecisionTreeClassifier(max_depth=1).fit(
X=train[feature_columns],
y=train[target_column]
)
train["pred_tree"] = tree.predict(train[feature_columns])
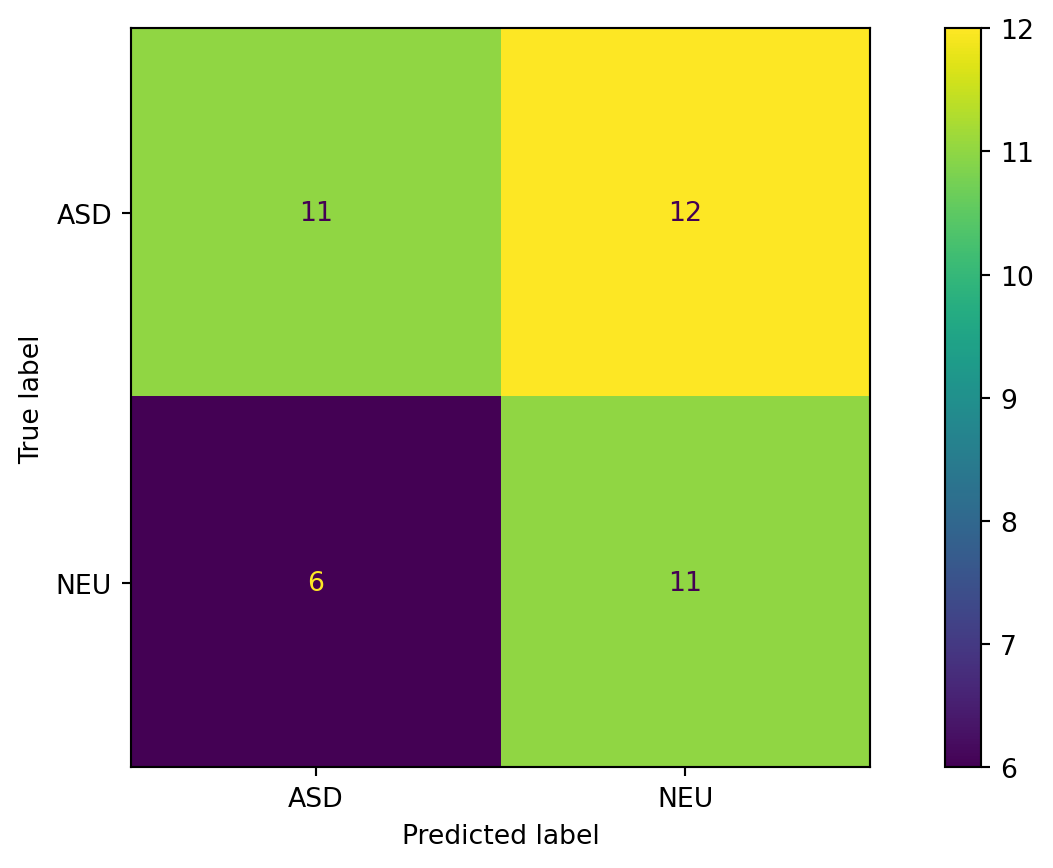
print("Training accuracy: ", accuracy_score(train[target_column], train["pred_tree"]))Training accuracy: 0.865546218487395Confusion Matrix
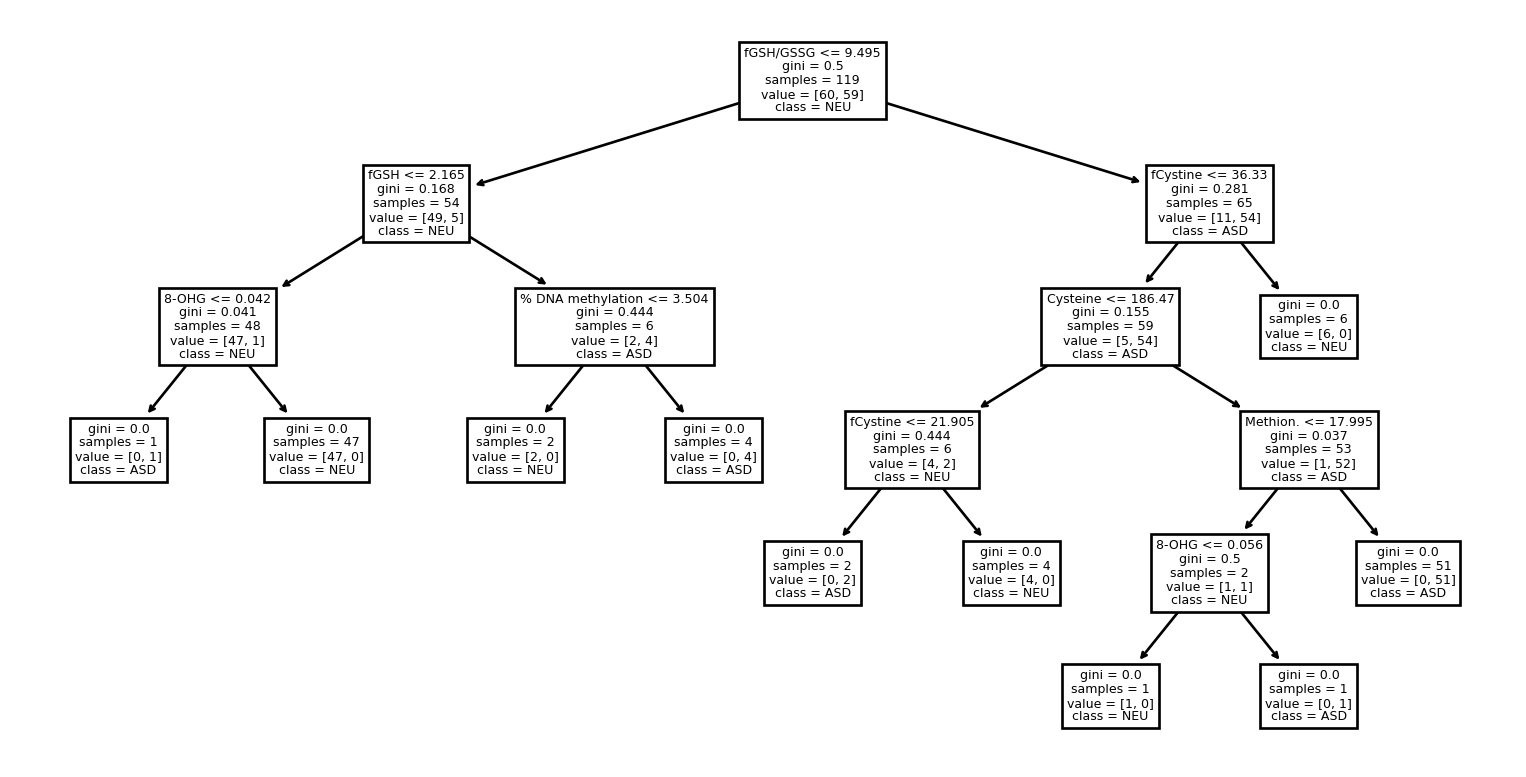
What does the tree look like?
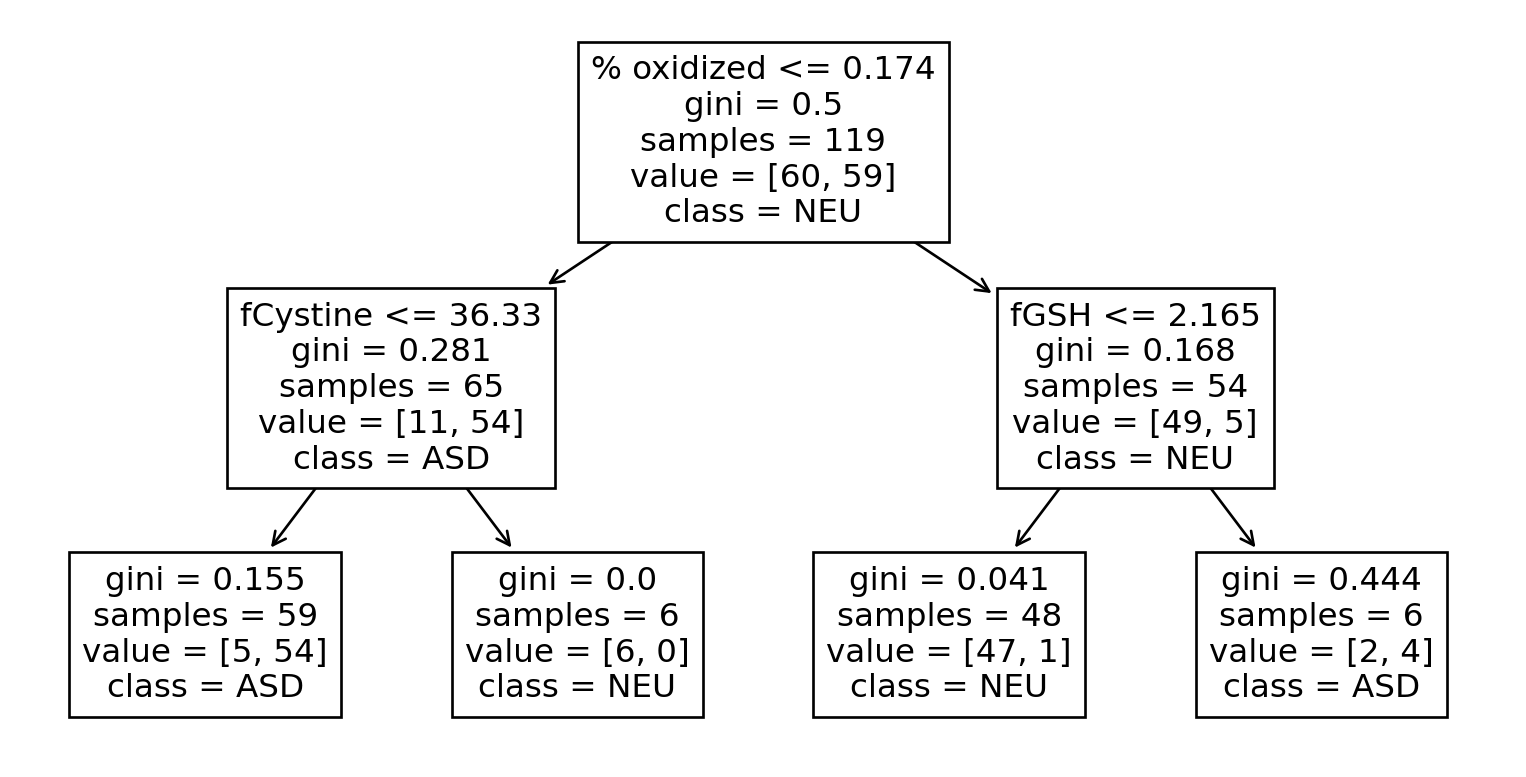
Go Deeper
Even Deeper
tree = DecisionTreeClassifier(max_depth=30).fit(
X=train[feature_columns],
y=train[target_column]
)
train["pred_tree"] = tree.predict(train[feature_columns])
print("Training accuracy: ", accuracy_score(train[target_column], train["pred_tree"]))Training accuracy: 1.0What do you think the test accuracy will be?
What decisions can be made at each node
- Continuous variables: compare one feature against a threshold value; if comparison is true go right, else go left
- Categorical variables: if is one of a set of categories, go right, else left
- At leaf nodes:
- regression tree: compute the mean value of items there, predict that.
- classification tree: compute the proportion of each category for items there
- predict the most common category
- or: predict that unseen items will follow the same proportions
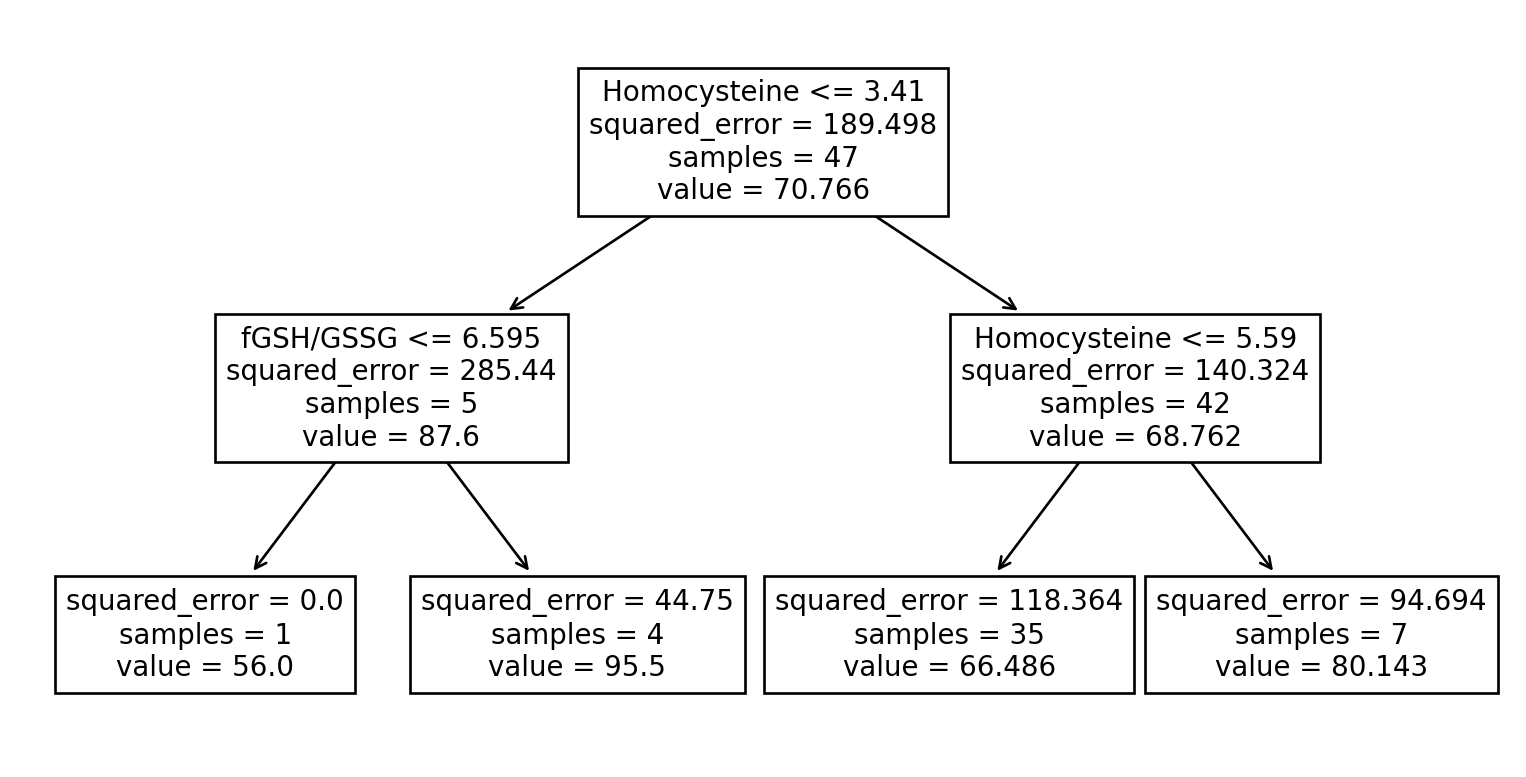
Example of a Regression Tree
We’ll try predicting the “Vineland ABC” score from the metabolites (for just the ASD children).
We’ll skip the usual train-test split because we’re just showing what the tree looks like.
What a regression tree looks like
Activity
Training Data (Classification Tree)
Dataset 0
| A | D | E |
|---|---|---|
| 1734 | Attchd | No |
| 1422 | Attchd | Yes |
| 1464 | Attchd | Yes |
| 2320 | Detchd | Yes |
| 2290 | Detchd | No |
| 1969 | Detchd | No |
Dataset 1
| B | C | E |
|---|---|---|
| 1973 | Twnhs | No |
| 1980 | Duplex | No |
| 2002 | OneFam | Yes |
| 1962 | OneFam | No |
| 1994 | OneFam | Yes |
| 1994 | OneFam | Yes |
Dataset 2
| A | C | E |
|---|---|---|
| 1367 | TwnhsE | Yes |
| 1512 | OneFam | Yes |
| 1149 | OneFam | No |
| 796 | OneFam | No |
| 1264 | OneFam | Yes |
| 1314 | OneFam | No |
Dataset 3
| A | C | E |
|---|---|---|
| 932 | OneFam | No |
| 1242 | OneFam | Yes |
| 1668 | OneFam | No |
| 1092 | TwoFmCon | No |
| 1226 | TwnhsE | Yes |
| 2418 | OneFam | Yes |
Dataset 4
| A | C | E |
|---|---|---|
| 864 | OneFam | No |
| 1434 | OneFam | No |
| 1196 | OneFam | No |
| 1720 | OneFam | Yes |
| 2787 | Duplex | Yes |
| 1586 | Twnhs | Yes |
Dataset 5
| B | D | E |
|---|---|---|
| 1900 | Detchd | No |
| 2006 | Attchd | Yes |
| 1929 | Detchd | Yes |
| 1940 | Detchd | No |
| 1970 | Attchd | Yes |
| 1916 | Detchd | No |
Dataset 6
| B | C | E |
|---|---|---|
| 2000 | OneFam | Yes |
| 1967 | OneFam | No |
| 1974 | OneFam | Yes |
| 1997 | OneFam | Yes |
| 1956 | OneFam | No |
| 1948 | OneFam | No |
Dataset 7
| B | C | E |
|---|---|---|
| 2005 | OneFam | Yes |
| 1950 | OneFam | Yes |
| 1937 | OneFam | No |
| 1915 | OneFam | No |
| 1980 | OneFam | Yes |
| 1971 | OneFam | No |
Dataset 8
| A | C | E |
|---|---|---|
| 1403 | OneFam | Yes |
| 1342 | OneFam | No |
| 2161 | OneFam | Yes |
| 1092 | Twnhs | No |
| 1852 | OneFam | Yes |
| 1578 | OneFam | No |
Dataset 9
| B | C | E |
|---|---|---|
| 1959 | OneFam | Yes |
| 1914 | OneFam | No |
| 1931 | OneFam | Yes |
| 2001 | OneFam | Yes |
| 1930 | OneFam | No |
| 1936 | OneFam | No |
Test set
| A | B | C | D |
|---|---|---|---|
| 1502 | 1923 | OneFam | Detchd |
| 1561 | 1960 | OneFam | Attchd |
| 2650 | 1967 | Duplex | Other |
| 1328 | 1959 | OneFam | Attchd |
| 3228 | 1992 | OneFam | Attchd |
| 1774 | 1900 | OneFam | Other |
Training Data (Regression Tree)
Dataset 0
| A | D | F |
|---|---|---|
| 1734 | Attchd | 126.0 |
| 1422 | Attchd | 179.6 |
| 1464 | Attchd | 282.9 |
| 2320 | Detchd | 259.5 |
| 2290 | Detchd | 122.5 |
| 1969 | Detchd | 141.0 |
Dataset 1
| B | C | F |
|---|---|---|
| 1973 | Twnhs | 119.5 |
| 1980 | Duplex | 144.0 |
| 2002 | OneFam | 220.0 |
| 1962 | OneFam | 130.0 |
| 1994 | OneFam | 193.5 |
| 1994 | OneFam | 301.5 |
Dataset 2
| A | C | F |
|---|---|---|
| 1367 | TwnhsE | 192.0 |
| 1512 | OneFam | 231.0 |
| 1149 | OneFam | 127.0 |
| 796 | OneFam | 85.0 |
| 1264 | OneFam | 167.5 |
| 1314 | OneFam | 145.0 |
Dataset 3
| A | C | F |
|---|---|---|
| 932 | OneFam | 124.0 |
| 1242 | OneFam | 175.5 |
| 1668 | OneFam | 135.0 |
| 1092 | TwoFmCon | 55.0 |
| 1226 | TwnhsE | 211.5 |
| 2418 | OneFam | 341.0 |
Dataset 4
| A | C | F |
|---|---|---|
| 864 | OneFam | 133.5 |
| 1434 | OneFam | 157.0 |
| 1196 | OneFam | 128.0 |
| 1720 | OneFam | 188.0 |
| 2787 | Duplex | 269.5 |
| 1586 | Twnhs | 170.0 |
Dataset 5
| B | D | F |
|---|---|---|
| 1900 | Detchd | 114.0 |
| 2006 | Attchd | 325.0 |
| 1929 | Detchd | 230.0 |
| 1940 | Detchd | 155.0 |
| 1970 | Attchd | 240.1 |
| 1916 | Detchd | 135.0 |
Dataset 6
| B | C | F |
|---|---|---|
| 2000 | OneFam | 327.0 |
| 1967 | OneFam | 134.5 |
| 1974 | OneFam | 260.0 |
| 1997 | OneFam | 210.0 |
| 1956 | OneFam | 131.0 |
| 1948 | OneFam | 138.0 |
Dataset 7
| B | C | F |
|---|---|---|
| 2005 | OneFam | 415.0 |
| 1950 | OneFam | 257.0 |
| 1937 | OneFam | 119.5 |
| 1915 | OneFam | 123.0 |
| 1980 | OneFam | 204.0 |
| 1971 | OneFam | 119.5 |
Dataset 8
| A | C | F |
|---|---|---|
| 1403 | OneFam | 202.0 |
| 1342 | OneFam | 105.0 |
| 2161 | OneFam | 230.5 |
| 1092 | Twnhs | 85.5 |
| 1852 | OneFam | 230.0 |
| 1578 | OneFam | 133.0 |
Dataset 9
| B | C | F |
|---|---|---|
| 1959 | OneFam | 200.0 |
| 1914 | OneFam | 67.0 |
| 1931 | OneFam | 169.5 |
| 2001 | OneFam | 421.2 |
| 1930 | OneFam | 110.0 |
| 1936 | OneFam | 115.0 |
Test set
| A | B | C | D |
|---|---|---|---|
| 1502 | 1923 | OneFam | Detchd |
| 1561 | 1960 | OneFam | Attchd |
| 2650 | 1967 | Duplex | Other |
| 1328 | 1959 | OneFam | Attchd |
| 3228 | 1992 | OneFam | Attchd |
| 1774 | 1900 | OneFam | Other |
Instructions
Columns A through D are features. Columns E and F are outcomes (targets).
- Pick a random number between 0 and 9 (inclusive). This is your dataset number.
- On paper, construct a decision tree to predict
Efrom the features in your dataset.- This doesn’t have to be the best possible tree; just try to come up with some reasonable tree.
- Make a second tree to predict
Ffrom the features in your dataset. - Compute the accuracy and MAE of your tree on your training set.
- Write down what your tree predicts for both
EandFfor each item in the test set.
Don’t peek beyond this slide until you’re done!
Evaluation
Here’s the test set with labels:
| Gr_Liv_Area | Year_Built | Bldg_Type | Garage_Type | sale_price_above_median | sale_price |
|---|---|---|---|---|---|
| 1502 | 1923 | OneFam | Detchd | Yes | 165.000000 |
| 1561 | 1960 | OneFam | Attchd | Yes | 193.000000 |
| 2650 | 1967 | Duplex | Other | Yes | 160.000000 |
| 1328 | 1959 | OneFam | Attchd | Yes | 170.000000 |
| 3228 | 1992 | OneFam | Attchd | Yes | 430.000000 |
| 1774 | 1900 | OneFam | Other | No | 87.000000 |
Compute your accuracy and MAE.
Reference Results
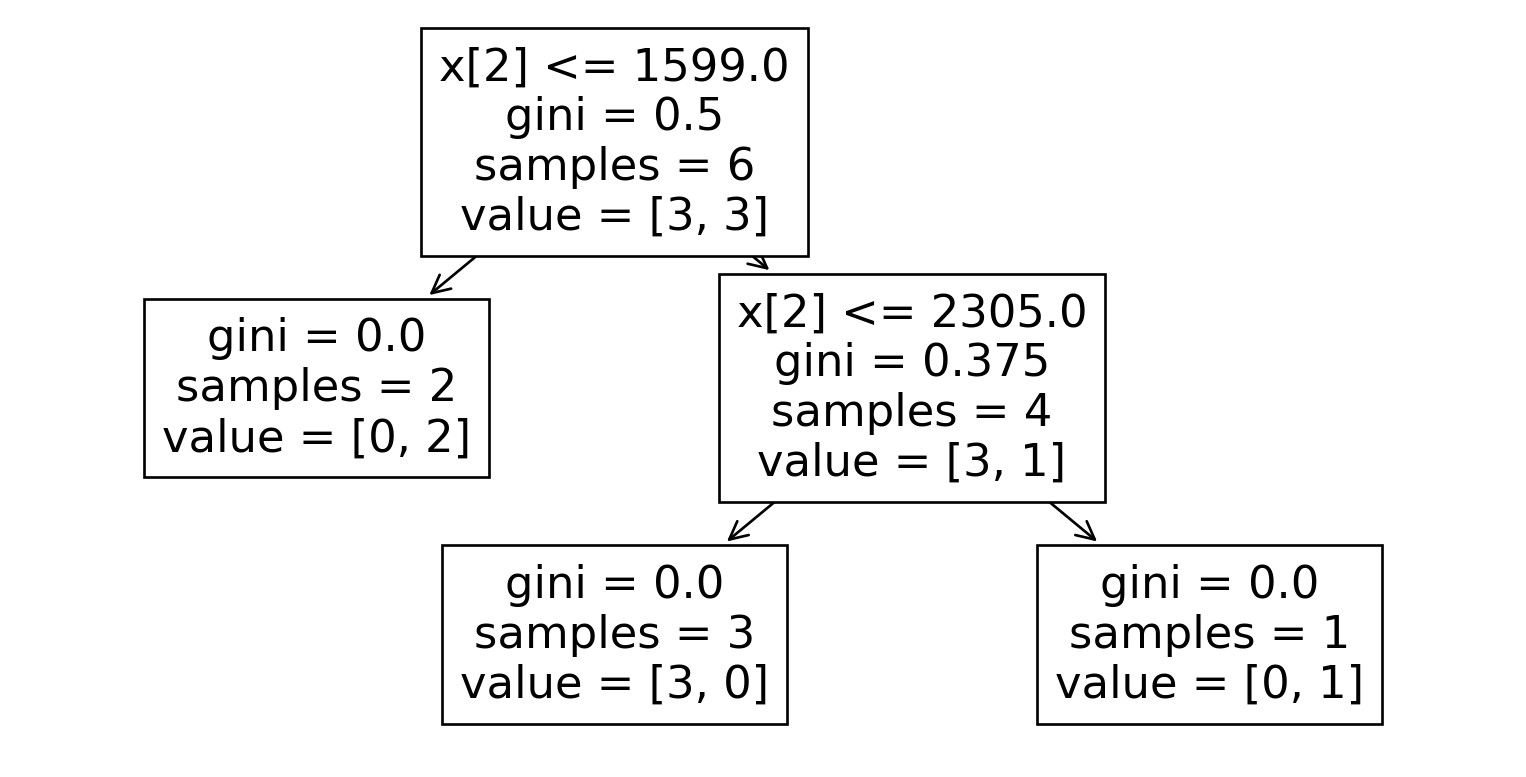
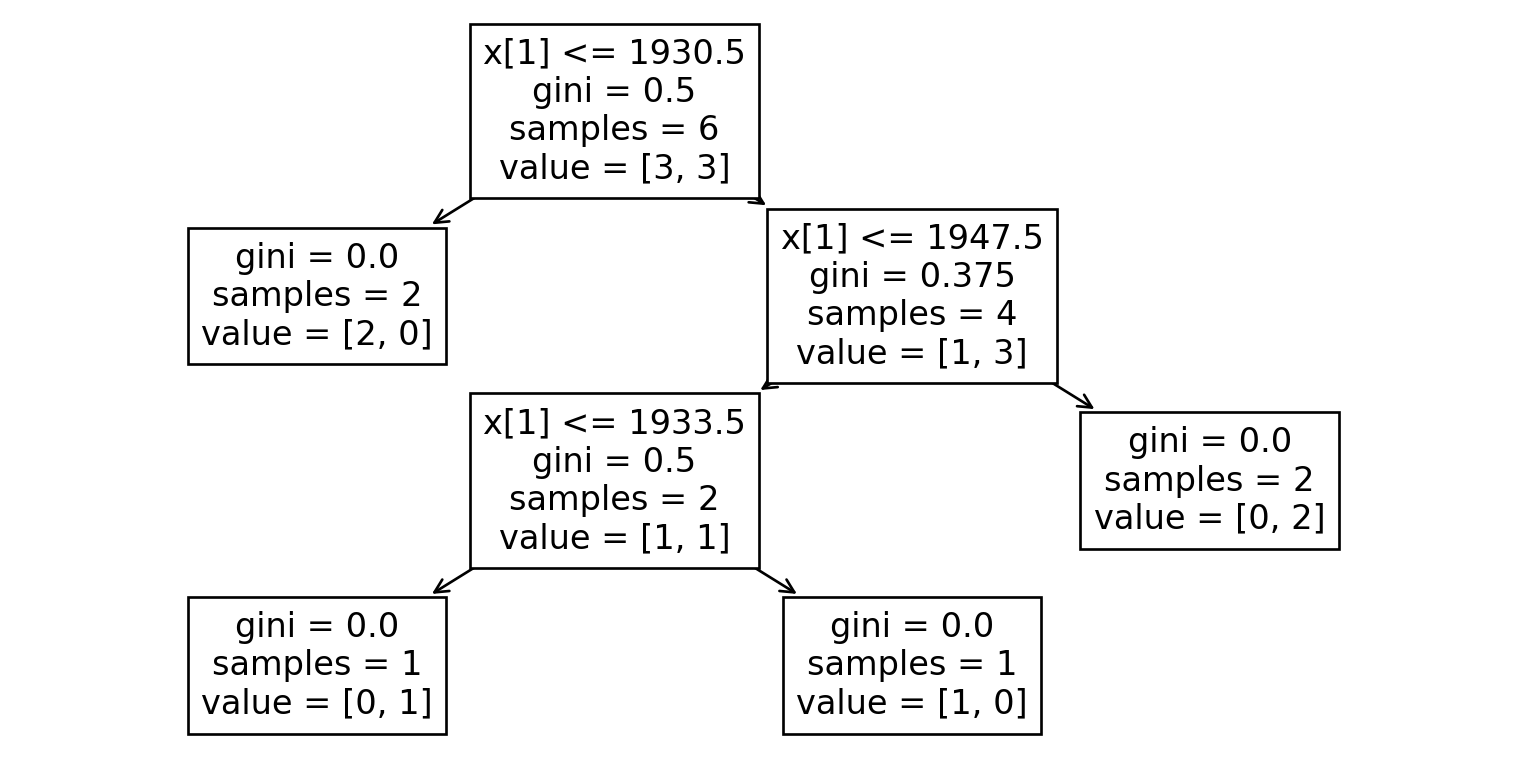
Here’s what classification trees look like for each dataset:
Dataset 0
| A | D | E |
|---|---|---|
| 1734 | Attchd | No |
| 1422 | Attchd | Yes |
| 1464 | Attchd | Yes |
| 2320 | Detchd | Yes |
| 2290 | Detchd | No |
| 1969 | Detchd | No |
Dataset 1
| B | C | E |
|---|---|---|
| 1973 | Twnhs | No |
| 1980 | Duplex | No |
| 2002 | OneFam | Yes |
| 1962 | OneFam | No |
| 1994 | OneFam | Yes |
| 1994 | OneFam | Yes |
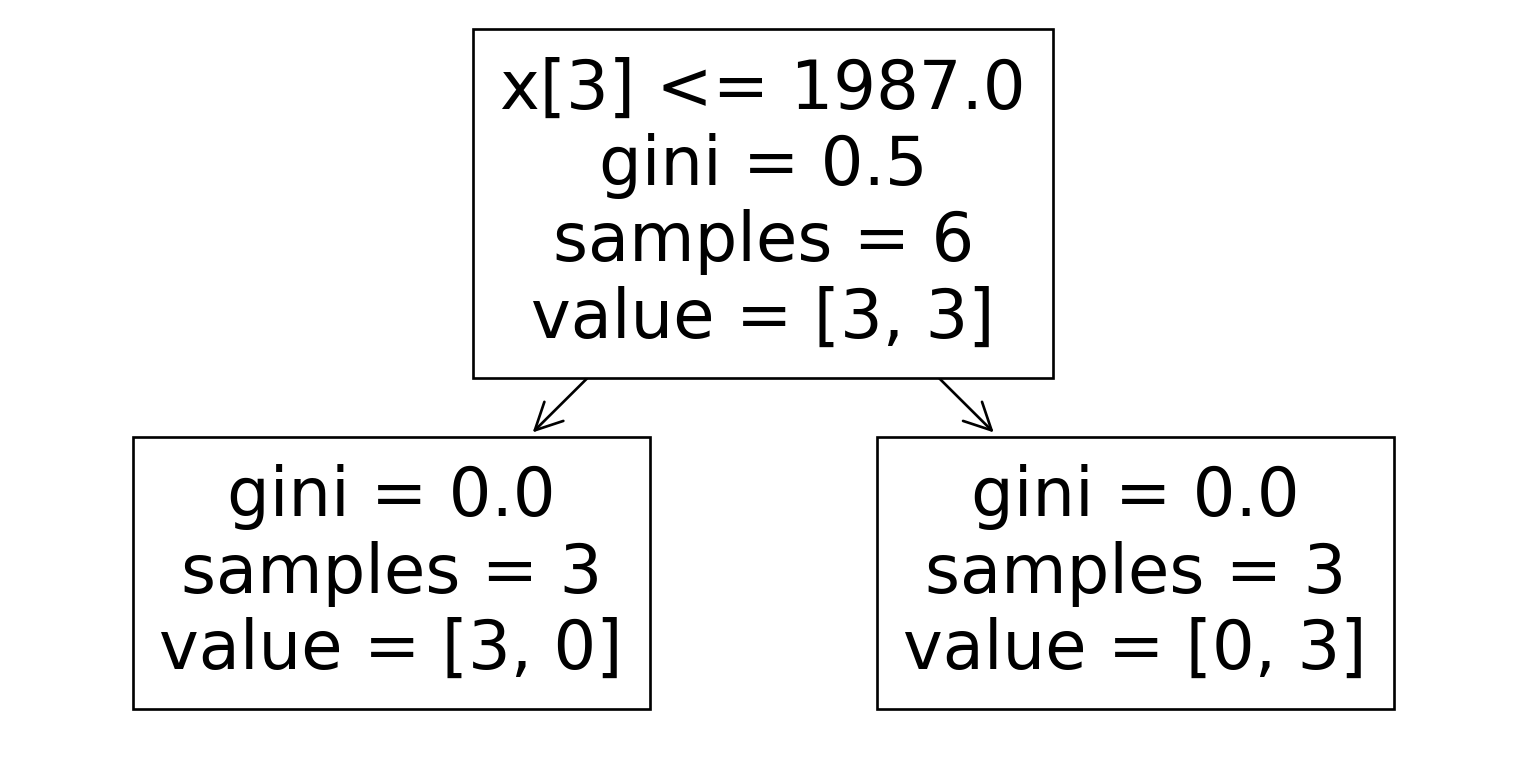
Dataset 2
| A | C | E |
|---|---|---|
| 1367 | TwnhsE | Yes |
| 1512 | OneFam | Yes |
| 1149 | OneFam | No |
| 796 | OneFam | No |
| 1264 | OneFam | Yes |
| 1314 | OneFam | No |
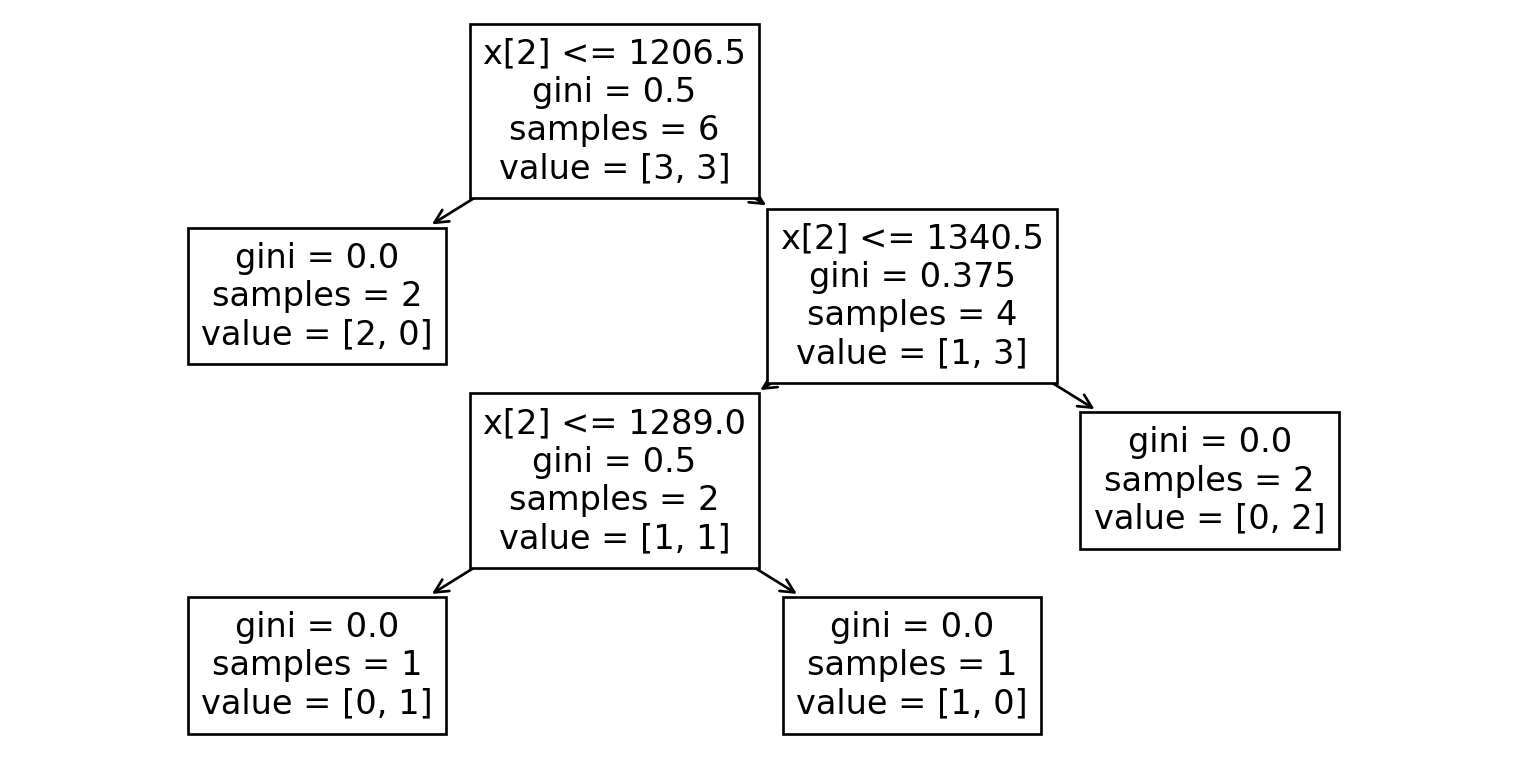
Dataset 3
| A | C | E |
|---|---|---|
| 932 | OneFam | No |
| 1242 | OneFam | Yes |
| 1668 | OneFam | No |
| 1092 | TwoFmCon | No |
| 1226 | TwnhsE | Yes |
| 2418 | OneFam | Yes |
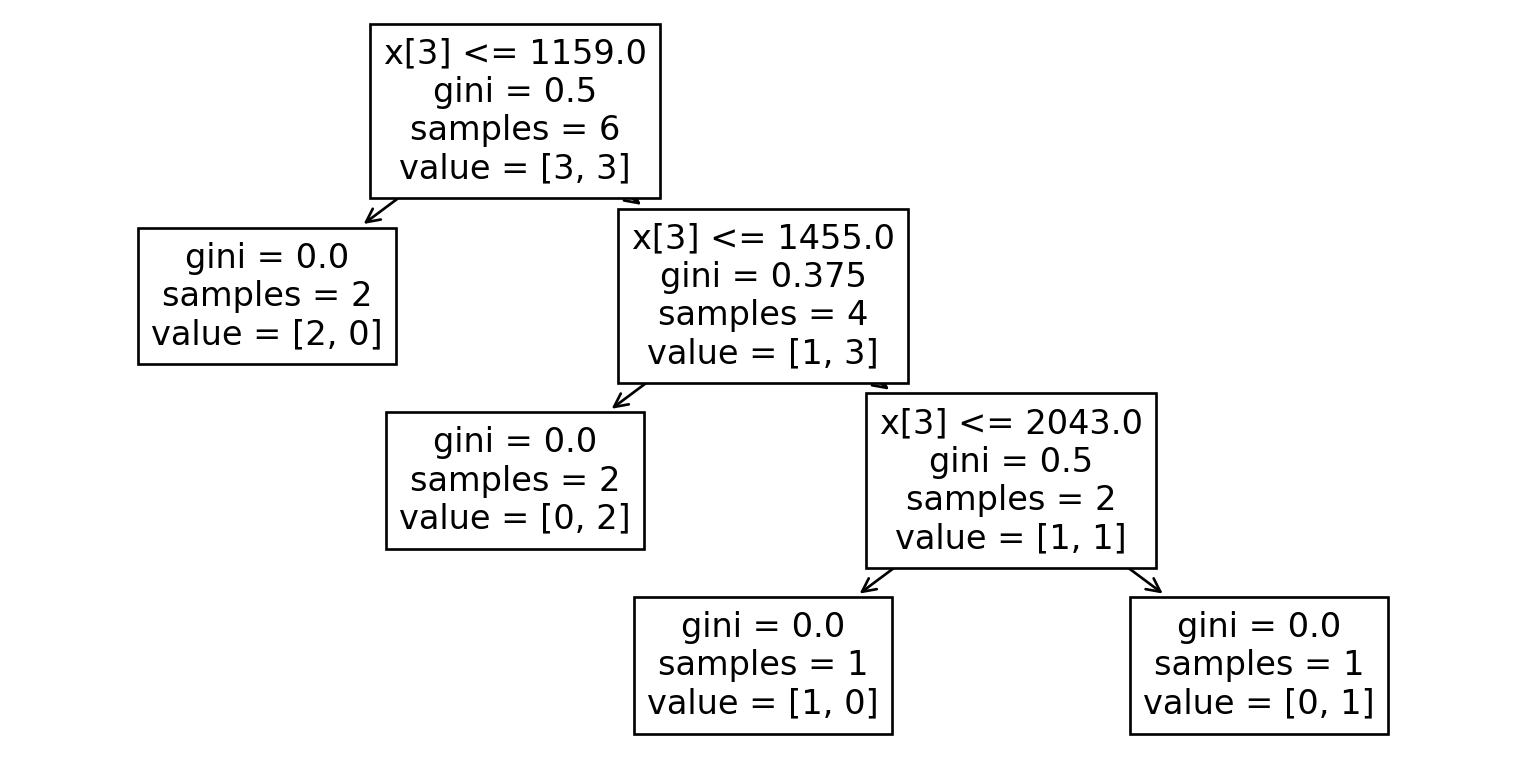
Dataset 4
| A | C | E |
|---|---|---|
| 864 | OneFam | No |
| 1434 | OneFam | No |
| 1196 | OneFam | No |
| 1720 | OneFam | Yes |
| 2787 | Duplex | Yes |
| 1586 | Twnhs | Yes |
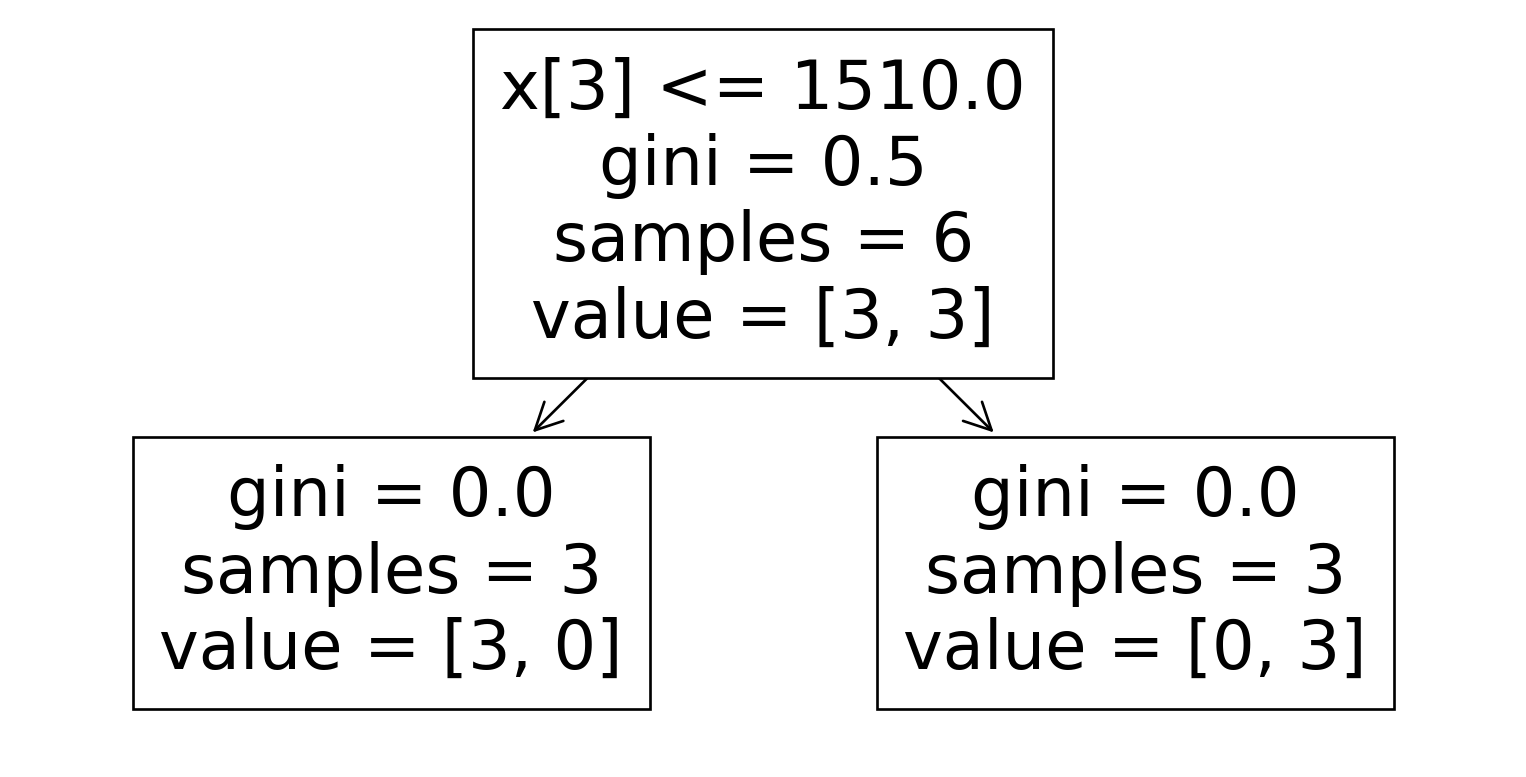
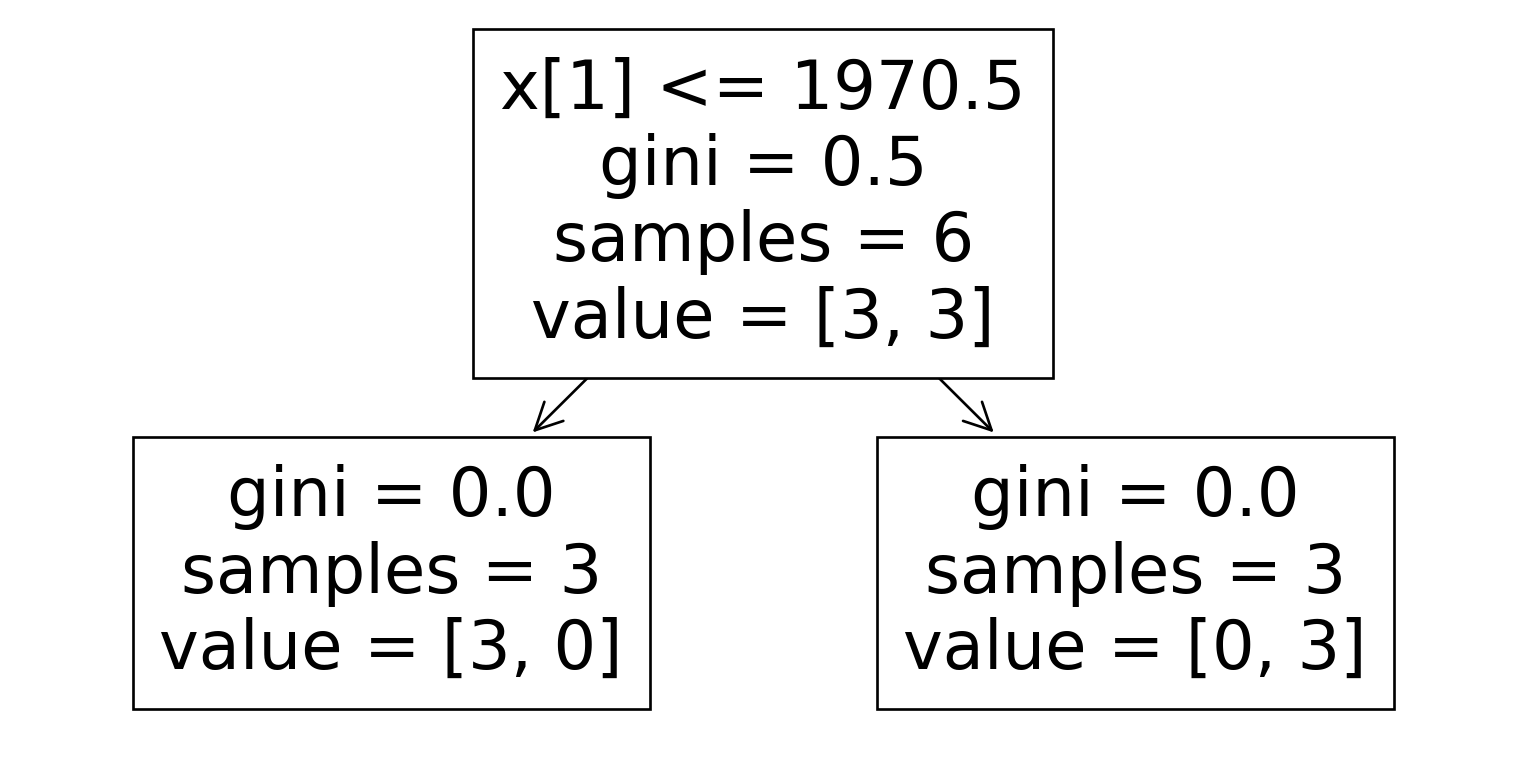
Dataset 5
| B | D | E |
|---|---|---|
| 1900 | Detchd | No |
| 2006 | Attchd | Yes |
| 1929 | Detchd | Yes |
| 1940 | Detchd | No |
| 1970 | Attchd | Yes |
| 1916 | Detchd | No |
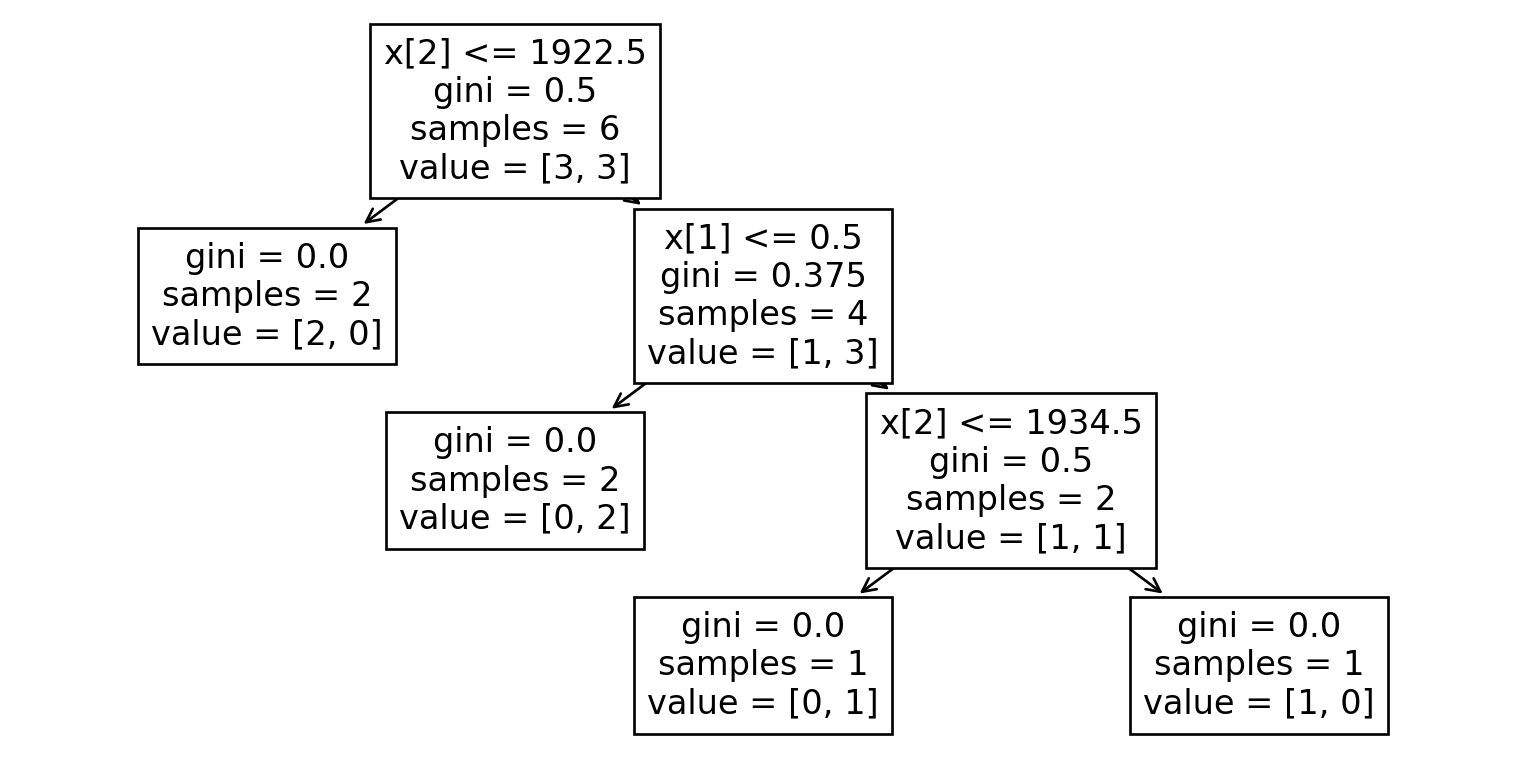
Dataset 6
| B | C | E |
|---|---|---|
| 2000 | OneFam | Yes |
| 1967 | OneFam | No |
| 1974 | OneFam | Yes |
| 1997 | OneFam | Yes |
| 1956 | OneFam | No |
| 1948 | OneFam | No |
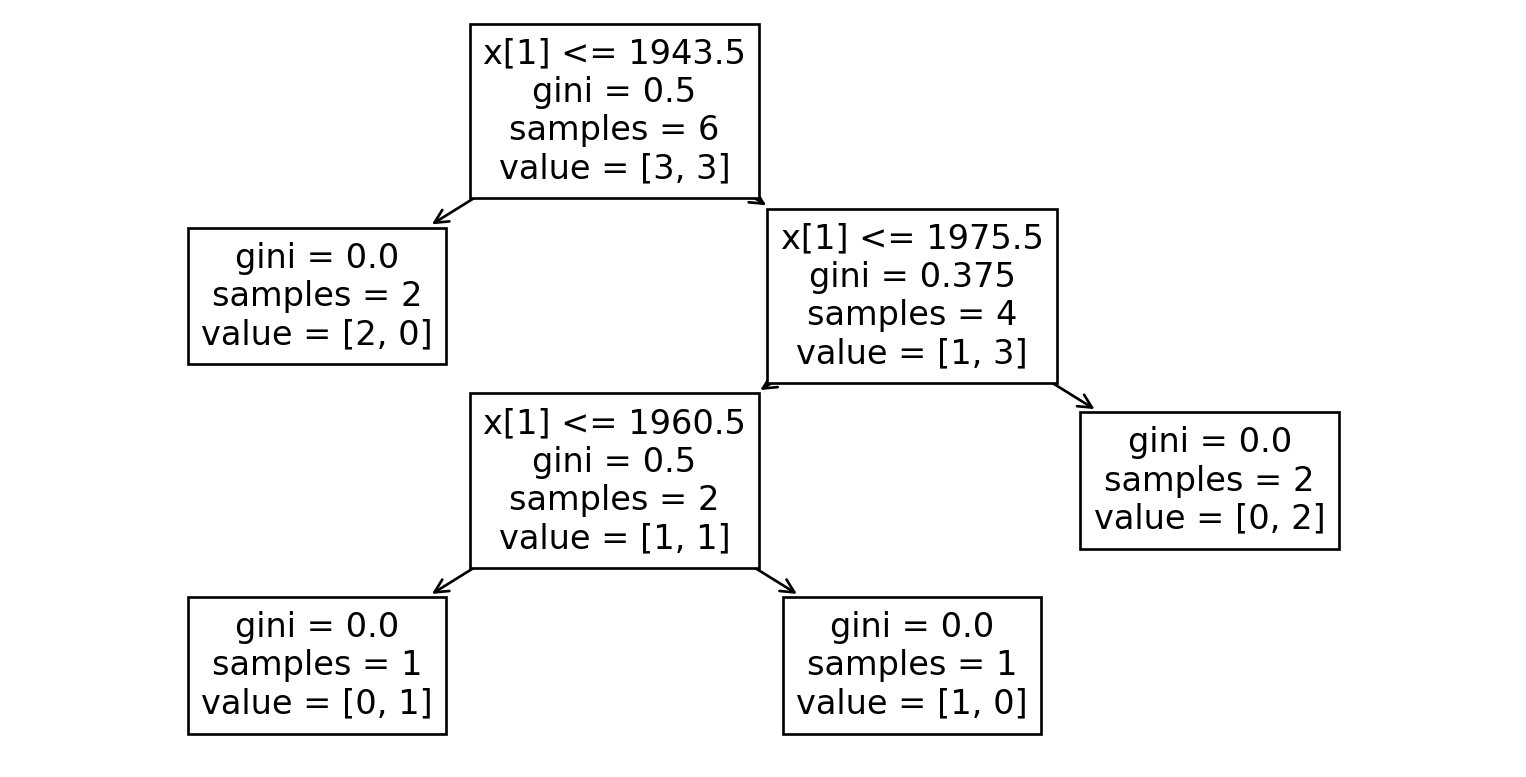
Dataset 7
| B | C | E |
|---|---|---|
| 2005 | OneFam | Yes |
| 1950 | OneFam | Yes |
| 1937 | OneFam | No |
| 1915 | OneFam | No |
| 1980 | OneFam | Yes |
| 1971 | OneFam | No |
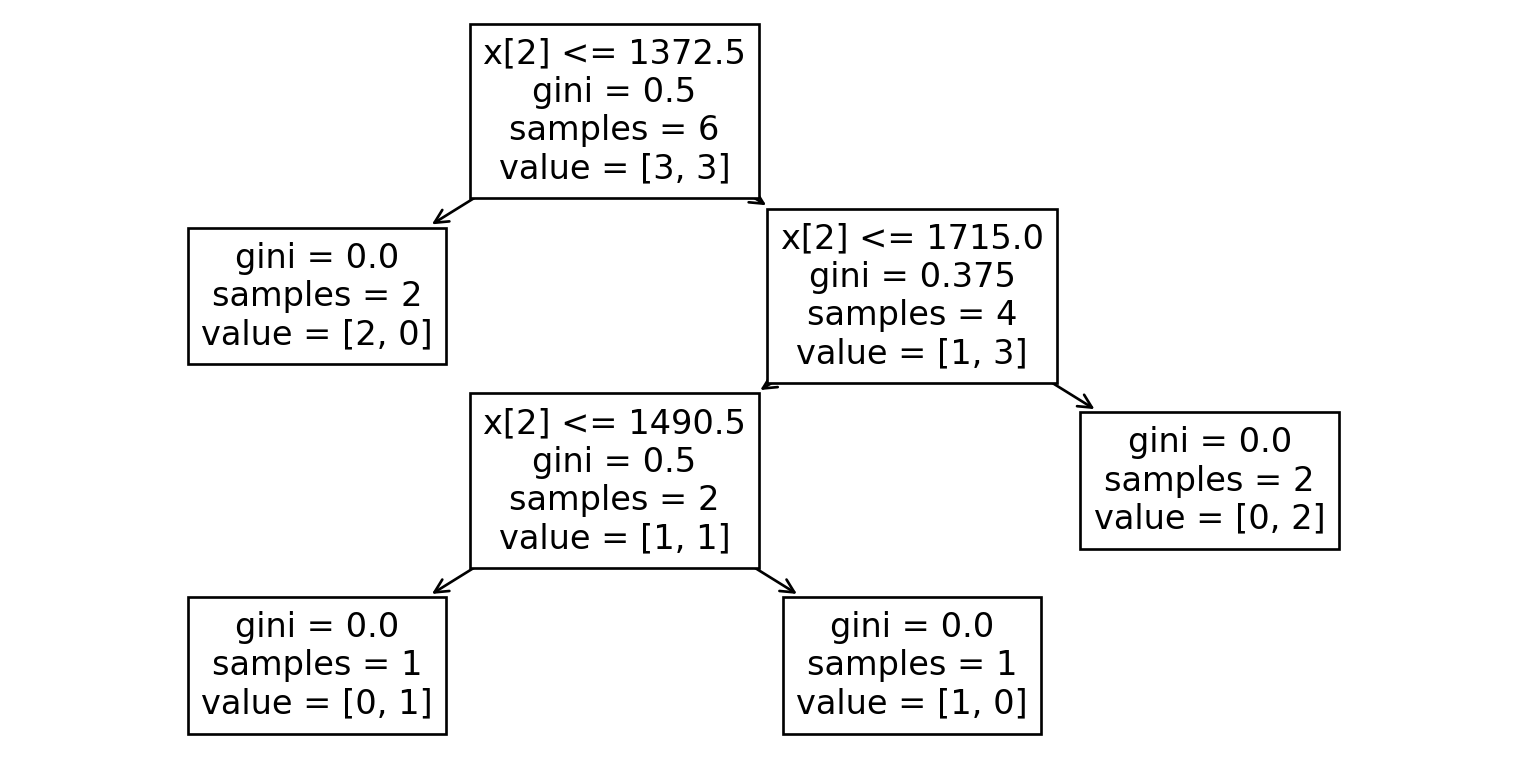
Dataset 8
| A | C | E |
|---|---|---|
| 1403 | OneFam | Yes |
| 1342 | OneFam | No |
| 2161 | OneFam | Yes |
| 1092 | Twnhs | No |
| 1852 | OneFam | Yes |
| 1578 | OneFam | No |
Dataset 9
| B | C | E |
|---|---|---|
| 1959 | OneFam | Yes |
| 1914 | OneFam | No |
| 1931 | OneFam | Yes |
| 2001 | OneFam | Yes |
| 1930 | OneFam | No |
| 1936 | OneFam | No |
Test set
| Gr_Liv_Area | Year_Built | Bldg_Type | Garage_Type | yes_votes | sale_price_above_median | Tree 1 | Tree 2 | Tree 3 | Tree 4 | Tree 5 | Tree 6 | Tree 7 | Tree 8 | Tree 9 | Tree 10 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1502 | 1923 | OneFam | Detchd | 0.300000 | Yes | Yes | No | Yes | No | No | Yes | No | No | No | No |
| 1561 | 1960 | OneFam | Attchd | 0.600000 | Yes | Yes | No | Yes | No | Yes | Yes | No | Yes | No | Yes |
| 2650 | 1967 | Duplex | Other | 0.700000 | Yes | Yes | No | Yes | Yes | Yes | Yes | No | No | Yes | Yes |
| 1328 | 1959 | OneFam | Attchd | 0.500000 | Yes | Yes | No | No | Yes | No | Yes | No | Yes | No | Yes |
| 3228 | 1992 | OneFam | Attchd | 1.000000 | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| 1774 | 1900 | OneFam | Other | 0.300000 | No | No | No | Yes | No | Yes | No | No | No | Yes | No |